Expectation Maximization for Logistic Mixture Autoregressive (LMAR) model. Attempt #2
Applying EM algorithm to regime-switching autoregressive processes.
Some time ago I wrote an article describing my unsuccessful attempt to implement Expectation Maximization algorithm for estimating parameters of Logistic Mixture Autoregressive (LMAR) model. In this article I am going to describe how I was able to fix the problem and provide a correct implementation of the algorithm. I will also provide code for estimating the order of the model and replicate some of the simulation studies from the paper the article is based on (‘Basket trading under co-integration with the logistic mixture autoregressive mode’ Cheng et al. 2011). I am not going to describe the model again here, please read the previous article to get familiar with it. Let’s start right away with the problem and how to fix it.
The implementation of EM algorithm in the previous article worked quite well when some of the parameters were treated as known. I performed several experiments treating different sets of parameters as known and in all of the cases was able to get good estimates of the true parameters. But when I tried estimating all of the parameters together, everything fell apart. All estimates were completely wrong.
I believe that the problem was that the model was not identifiable. There was no way for the EM algorithm to distinguish between the two regimes. Sometimes the first regime was determined to be stationary (which is correct) and sometimes the second regime was stationary (which is wrong). In some simulations probably both regimes were stationary.
To fix this problem a simple convergence condition was needed — condition that the second regime is non-stationary. It is done with a couple lines of code.
The parameters of the model used along with the code to generate and plot a sample from it are shown below.
| N = 500 # number of datapoints to generate | |
| ksi = [-1.3, 0.6, 0.3] | |
| zeta_1 = [0, 0.6, -0.2] | |
| var_1 = 1 | |
| zeta_2 = [0, 1.5] | |
| var_2 = 3 | |
| np.random.seed(5) | |
| a = generate_a(N, ksi, zeta_1, var_1, zeta_2, var_2) | |
| plt.figure(figsize=(18,6)) | |
| plt.plot(a) |
As can be seen from parameter vector zeta_2, the second regime is AR(1) model. For AR(1) model to be non-stationary its lag coefficient must be greater than or equal to 1. So that’s what we must check for.
The new implementation of EM algorithm is shown below. The only difference is on lines 96–97 where we check that the second regime is non-stationary. If this condition is not satisfied, the algorithm is restarted with new initial parameters.
| def em_algorithm(a, max_iter, tol): | |
| ''' | |
| run em algorithm for max_iter iterations or until convergence | |
| ''' | |
| npdf = stats.norm.pdf | |
| converged = False | |
| while not converged: | |
| try: | |
| # random initial guess | |
| ksi = np.random.uniform(-3,3,(3)) | |
| zeta_1 = np.random.uniform(-3,3,(3)) | |
| zeta_2 = np.random.uniform(-3,3,(2)) | |
| var_1 = np.random.uniform(0,6,(1)) | |
| var_2 = np.random.uniform(0,6,(1)) | |
| # create dataframe | |
| df = pd.DataFrame(columns=['const', 'a', 'a_l1', 'a_l2', 'abs_l1', 'abs_l2', 'p1', 'p2', | |
| 'tau1', 'tau2', 'e1', 'e2']) | |
| df['a'] = a[2:] | |
| df['a_l1'] = a[1:-1] | |
| df['a_l2'] = a[:-2] | |
| df['abs_l1'] = abs(df['a_l1']) | |
| df['abs_l2'] = abs(df['a_l2']) | |
| df['const'] = 1 | |
| for s in range(max_iter): | |
| # E-step | |
| # solve for mixing probabilities | |
| num = np.exp((df[['const', 'abs_l1', 'abs_l2']] * ksi).sum(axis=1)) | |
| denom = (1 + np.exp((df[['const', 'abs_l1', 'abs_l2']] * ksi).sum(axis=1))) | |
| df['p1'] = num/denom | |
| df['p2'] = 1 - df['p1'] | |
| # calculate e | |
| df['e1'] = df['a'] - (zeta_1 * df[['const', 'a_l1', 'a_l2']]).sum(axis=1) | |
| df['e2'] = df['a'] - (zeta_2 * df[['const', 'a_l1']]).sum(axis=1) | |
| # calculate tau | |
| sigma_1 = np.sqrt(var_1) | |
| sigma_2 = np.sqrt(var_2) | |
| denom = ((df['p1'] / sigma_1) * stats.norm.pdf(df['e1'] / sigma_1) + | |
| (df['p2'] / sigma_2) * stats.norm.pdf(df['e2'] / sigma_2)) | |
| df['tau1'] = (df['p1'] / sigma_1) * stats.norm.pdf(df['e1'] / sigma_1) / denom | |
| df['tau2'] = (df['p2'] / sigma_2) * stats.norm.pdf(df['e2'] / sigma_2) / denom | |
| # M-step | |
| # compute ksi | |
| dr_dksi = df[['const', 'abs_l1', 'abs_l2']] | |
| dQ_dksi = (np.tile((df['tau1'] - df['p1']), (3,1)).T * dr_dksi).sum().values | |
| df['dr_dksi^2'] = df[['const', 'abs_l1', 'abs_l2']].apply(outer, axis=1).apply(np.array) | |
| d2Q_dksi2 = -(df['p1'] * df['p2'] * df['dr_dksi^2']).sum() | |
| new_ksi = ksi - np.linalg.inv(d2Q_dksi2) @ dQ_dksi | |
| # compute zeta | |
| df['de1_dzeta1^2'] = df[['const', 'a_l1', 'a_l2']].apply(outer, axis=1).apply(np.array) | |
| df['de2_dzeta2^2'] = df[['const', 'a_l1']].apply(outer, axis=1).apply(np.array) | |
| zeta1_sum1 = (df['tau1'] * df['de1_dzeta1^2']).sum() | |
| zeta2_sum1 = (df['tau2'] * df['de2_dzeta2^2']).sum() | |
| zeta1_sum2 = (-np.tile(df['tau1'] * df['a'], (3,1)).T * -df[['const', 'a_l1', 'a_l2']]).sum().values | |
| zeta2_sum2 = (-np.tile(df['tau2'] * df['a'], (2,1)).T * -df[['const', 'a_l1']]).sum().values | |
| new_zeta_1 = np.linalg.inv(zeta1_sum1) @ zeta1_sum2 | |
| new_zeta_2 = np.linalg.inv(zeta2_sum1) @ zeta2_sum2 | |
| # compute variance | |
| sigma1_sum2 = (df['tau1'] * (df['a'] + np.dot(-df[['const', 'a_l1', 'a_l2']], new_zeta_1))**2).sum() | |
| sigma2_sum2 = (df['tau2'] * (df['a'] + np.dot(-df[['const', 'a_l1']], new_zeta_2))**2).sum() | |
| new_var_1 = 1/df['tau1'].sum() * sigma1_sum2 | |
| new_var_2 = 1/df['tau2'].sum() * sigma2_sum2 | |
| # calculate the difference between old and new values of parameters | |
| delta_ksi = np.sum(abs(new_ksi - ksi)) | |
| delta_zeta_1 = np.sum(abs(new_zeta_1 - zeta_1)) | |
| delta_zeta_2 = np.sum(abs(new_zeta_2 - zeta_2)) | |
| delta_var_1 = np.sum(abs(new_var_1 - var_1)) | |
| delta_var_2 = np.sum(abs(new_var_2 - var_2)) | |
| # update parameters | |
| ksi = new_ksi | |
| zeta_1 = new_zeta_1 | |
| zeta_2 = new_zeta_2 | |
| var_1 = new_var_1 | |
| var_2 = new_var_2 | |
| # calculate total difference between old and new parameters | |
| delta_total = delta_ksi + delta_zeta_1 + delta_zeta_2 + delta_var_1 + delta_var_2 | |
| # check convergence | |
| if delta_total < tol: | |
| # convergence conditions | |
| conv_cond = zeta_2[1] >= 1 # second regime is non-stationary | |
| if conv_cond: | |
| print('Converged at iteration ', s) | |
| converged = True | |
| break | |
| else: | |
| break | |
| # if any parameter is nan, stop | |
| nansum = (np.isnan(ksi).sum() + np.isnan(zeta_1).sum() + np.isnan(zeta_2).sum() + | |
| np.isnan(var_1).sum() + np.isnan(var_2).sum()) | |
| if nansum != 0: | |
| break | |
| if not converged: | |
| print(f'Algorithm not converged') | |
| print('Repeating with different initital values') | |
| except: | |
| continue | |
| return ksi, zeta_1, var_1, zeta_2, var_2 |
To check that the algorithm works correctly we can perform some simulation studies. I am not going to perform exactly the same studies as in the paper because it takes too much time. I will use less simulations and a bigger sample size in each simulation.
| delta_0s = [] | |
| delta_1s = [] | |
| delta_2s = [] | |
| phi_10s = [] | |
| phi_11s = [] | |
| phi_12s = [] | |
| phi_20s = [] | |
| phi_21s = [] | |
| var_1s = [] | |
| var_2s = [] | |
| max_iter=50 | |
| tol = 0.05 | |
| N = 1000 # number of datapoints to generate | |
| N_sim = 100 # number of simulations to run | |
| # parameters used to generate a | |
| ksi = [-1.3, 0.6, 0.3] | |
| zeta_1 = [0, 0.6, -0.2] | |
| var_1 = 1 | |
| zeta_2 = [0, 1.5] | |
| var_2 = 3 | |
| for s in range(N_sim): | |
| print() | |
| print('Running sim ', s) | |
| # generate a | |
| np.random.seed(123+s) | |
| a = generate_a(N, ksi, zeta_1, var_1, zeta_2, var_2) | |
| # run EM algorithm | |
| ksi_hat, zeta_1_hat, var_1_hat, zeta_2_hat, var_2_hat = em_algorithm(a, max_iter, tol) | |
| # save results | |
| delta_0s.append(ksi_hat[0]) | |
| delta_1s.append(ksi_hat[1]) | |
| delta_2s.append(ksi_hat[2]) | |
| phi_10s.append(zeta_1_hat[0]) | |
| phi_11s.append(zeta_1_hat[1]) | |
| phi_12s.append(zeta_1_hat[2]) | |
| phi_20s.append(zeta_2_hat[0]) | |
| phi_21s.append(zeta_2_hat[1]) | |
| var_1s.append(var_1_hat) | |
| var_2s.append(var_2_hat) |
The results of each simulation can be combined in a dataframe for further study.
| res = pd.DataFrame.from_dict({r'$\sigma_1^2$':var_1s, r'$\phi_{1,0}$':phi_10s, r'$\phi_{1,1}$':phi_11s, | |
| r'$\phi_{1,2}$':phi_12s, r'$\sigma_2^2$':var_2s, r'$\phi_{2,0}$':phi_20s, | |
| r'$\phi_{2,1}$':phi_21s, r'$\delta_0$':delta_0s, r'$\delta_1$':delta_1s, | |
| r'$\delta_2$':delta_2s}) |
Let’s look at the first several rows of the dataframe.
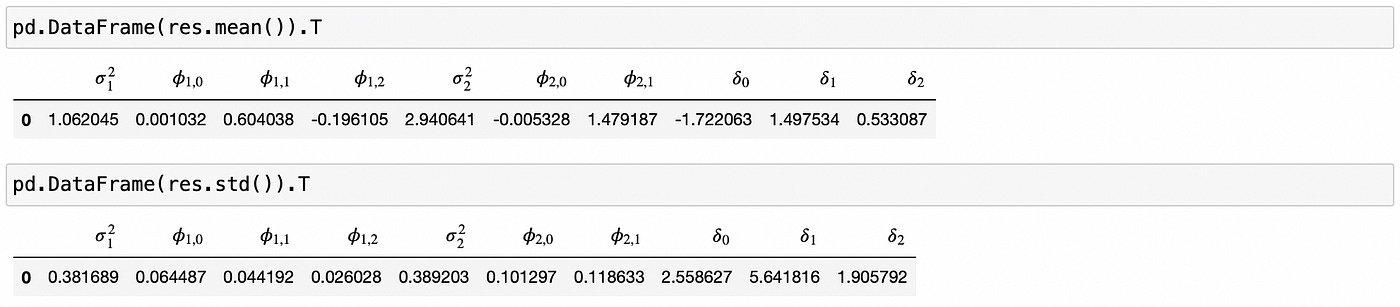
It seems that the estimates are more or less correct. At least in the ballpark. Now let’s look at the mean and standard deviation of all 100 simulations.
Here we can see some problems with parameters delta. The means are far from true values and standard deviations are too high. What’s going on here? Let’s look at the results in a little more detail.
If we look at the min and max statistics, we see that parameters delta have outliers that are very different from the estimates from most other simulations. In fact, if we just use median instead of mean as parameter estimates, we get numbers that are really close to true values.
So I think that this implementation of EM algorithm does sufficiently good job at estimating model parameters.
Now we need to generalize it to work with a model of any order (m1,m2,n). Code for it is demonstrated below. Most of the differences are minor and can be easily understood from the code. One important thing to note is how convergence condition is changed. Recall that we need to check whether the second regime is non-stationary. In the case of AR(1) model we did that by checking the lag coefficient. For a general case of AR(p) model we need to compute the reciprocal roots of characteristic polynomial and check that at least one of them is larger than or equal to one. It is done on line 110–117.
| def em_algorithm(a, m1, m2, n, max_iter, tol): | |
| ''' | |
| run em algorithm for max_iter iterations or until convergence | |
| a: time series | |
| m1: order of the first (stationary) AR model | |
| m2: order of the second (non-stationary) AR model | |
| n: number of lags in logistic equation | |
| ''' | |
| npdf = stats.norm.pdf | |
| max_lag = max(m1,m2,n) | |
| converged = False | |
| while not converged: | |
| #try: | |
| # random initial guess | |
| ksi = np.random.uniform(-3,3,(n+1)) | |
| zeta_1 = np.random.uniform(-3,3,(m1+1)) | |
| zeta_2 = np.random.uniform(-3,3,(m2+1)) | |
| var_1 = np.random.uniform(0,6,(1)) | |
| var_2 = np.random.uniform(0,6,(1)) | |
| # create dataframe | |
| a_lags = [f'a_l{i}' for i in range(1,max_lag+1)] | |
| abs_lags = [f'abs_l{i}' for i in range(1,n+1)] | |
| columns = ['const', 'a'] + a_lags + abs_lags + ['p1', 'p2', 'tau1', 'tau2', 'e1', 'e2'] | |
| df = pd.DataFrame(columns=columns) | |
| df['a'] = a[max_lag:] | |
| df['const'] = 1 | |
| for l in range(1,max_lag+1): | |
| df[a_lags[l-1]] = a[max_lag-l:-l] | |
| for l in range(1,n+1): | |
| df[abs_lags[l-1]] = abs(df[a_lags[l-1]]) | |
| for s in range(max_iter): | |
| # E-step | |
| # solve for mixing probabilities | |
| num = np.exp((df[['const'] + abs_lags] * ksi).sum(axis=1)) | |
| denom = (1 + np.exp((df[['const'] + abs_lags] * ksi).sum(axis=1))) | |
| df['p1'] = num/denom | |
| df['p2'] = 1 - df['p1'] | |
| # calculate e | |
| df['e1'] = df['a'] - (zeta_1 * df[['const'] + a_lags[:m1]]).sum(axis=1) | |
| df['e2'] = df['a'] - (zeta_2 * df[['const'] + a_lags[:m2]]).sum(axis=1) | |
| # calculate tau | |
| sigma_1 = np.sqrt(var_1) | |
| sigma_2 = np.sqrt(var_2) | |
| denom = ((df['p1'] / sigma_1) * stats.norm.pdf(df['e1'] / sigma_1) + | |
| (df['p2'] / sigma_2) * stats.norm.pdf(df['e2'] / sigma_2)) | |
| df['tau1'] = (df['p1'] / sigma_1) * stats.norm.pdf(df['e1'] / sigma_1) / denom | |
| df['tau2'] = (df['p2'] / sigma_2) * stats.norm.pdf(df['e2'] / sigma_2) / denom | |
| # M-step | |
| # compute ksi | |
| dr_dksi = df[['const'] + abs_lags] | |
| dQ_dksi = (np.tile((df['tau1'] - df['p1']), (n+1,1)).T * dr_dksi).sum().values | |
| df['dr_dksi^2'] = df[['const'] + abs_lags].apply(outer, axis=1).apply(np.array) | |
| d2Q_dksi2 = -(df['p1'] * df['p2'] * df['dr_dksi^2']).sum() | |
| new_ksi = ksi - np.linalg.inv(d2Q_dksi2) @ dQ_dksi | |
| # compute zeta | |
| df['de1_dzeta1^2'] = df[['const'] + a_lags[:m1]].apply(outer, axis=1).apply(np.array) | |
| df['de2_dzeta2^2'] = df[['const'] + a_lags[:m2]].apply(outer, axis=1).apply(np.array) | |
| zeta1_sum1 = (df['tau1'] * df['de1_dzeta1^2']).sum() | |
| zeta2_sum1 = (df['tau2'] * df['de2_dzeta2^2']).sum() | |
| zeta1_sum2 = (-np.tile(df['tau1'] * df['a'], (m1+1,1)).T * -df[['const'] + a_lags[:m1]]).sum().values | |
| zeta2_sum2 = (-np.tile(df['tau2'] * df['a'], (m2+1,1)).T * -df[['const'] + a_lags[:m2]]).sum().values | |
| new_zeta_1 = np.linalg.inv(zeta1_sum1) @ zeta1_sum2 | |
| new_zeta_2 = np.linalg.inv(zeta2_sum1) @ zeta2_sum2 | |
| # compute variance | |
| sigma1_sum2 = (df['tau1'] * (df['a'] + np.dot(-df[['const'] + a_lags[:m1]], new_zeta_1))**2).sum() | |
| sigma2_sum2 = (df['tau2'] * (df['a'] + np.dot(-df[['const'] + a_lags[:m2]], new_zeta_2))**2).sum() | |
| new_var_1 = 1/df['tau1'].sum() * sigma1_sum2 | |
| new_var_2 = 1/df['tau2'].sum() * sigma2_sum2 | |
| # calculate the difference between old and new values of parameters | |
| delta_ksi = np.sum(abs(new_ksi - ksi)) | |
| delta_zeta_1 = np.sum(abs(new_zeta_1 - zeta_1)) | |
| delta_zeta_2 = np.sum(abs(new_zeta_2 - zeta_2)) | |
| delta_var_1 = np.sum(abs(new_var_1 - var_1)) | |
| delta_var_2 = np.sum(abs(new_var_2 - var_2)) | |
| # update parameters | |
| ksi = new_ksi | |
| zeta_1 = new_zeta_1 | |
| zeta_2 = new_zeta_2 | |
| var_1 = new_var_1 | |
| var_2 = new_var_2 | |
| # if any parameter is nan, stop | |
| nansum = (np.isnan(ksi).sum() + np.isnan(zeta_1).sum() + np.isnan(zeta_2).sum() + | |
| np.isnan(var_1).sum() + np.isnan(var_2).sum()) | |
| if nansum != 0: | |
| break | |
| # calculate total difference between old and new parameters | |
| delta_total = delta_ksi + delta_zeta_1 + delta_zeta_2 + delta_var_1 + delta_var_2 | |
| # check convergence | |
| if delta_total < tol: | |
| # calculate roots of characteristic polynomial of the second regime | |
| char_pol = np.polynomial.polynomial.Polynomial(np.concatenate(([1],-zeta_2[1:]))) | |
| # calculate reciprocal roots and take absolute value | |
| roots = np.abs(1/char_pol.roots()) | |
| # convergence conditions | |
| # second regime is non-stationary | |
| conv_cond = sum(roots>=1) > 0 # at least one reciprocal root >= 1 | |
| if conv_cond: | |
| print('Converged at iteration ', s) | |
| converged = True | |
| break | |
| else: | |
| break | |
| if not converged: | |
| print(f'Algorithm not converged') | |
| print('Repeating with different initital values') | |
| #except: | |
| # continue | |
| return ksi, zeta_1, var_1, zeta_2, var_2 |
I will run a small simulation study just to check that the code works fine. I am going to use the same parameters as before, but run just 10 simulations. The only difference in the code is that now we need to pass the order of the model to em_algorithm function.
| delta_0s = [] | |
| delta_1s = [] | |
| delta_2s = [] | |
| phi_10s = [] | |
| phi_11s = [] | |
| phi_12s = [] | |
| phi_20s = [] | |
| phi_21s = [] | |
| var_1s = [] | |
| var_2s = [] | |
| max_iter=50 | |
| tol = 0.05 | |
| N = 1000 # number of datapoints to generate | |
| N_sim = 10 # number of simulations to run | |
| # assume the order of the model is known | |
| m1 = 2 | |
| m2 = 1 | |
| n = 2 | |
| # parameters used to generate a | |
| ksi = [-1.3, 0.6, 0.3] | |
| zeta_1 = [0, 0.6, -0.2] | |
| var_1 = 1 | |
| zeta_2 = [0, 1.5] | |
| var_2 = 3 | |
| for s in range(N_sim): | |
| print() | |
| print('Running sim ', s) | |
| # generate a | |
| np.random.seed(123+s) | |
| a = generate_a(N, ksi, zeta_1, var_1, zeta_2, var_2) | |
| # run EM algorithm | |
| ksi_hat, zeta_1_hat, var_1_hat, zeta_2_hat, var_2_hat = em_algorithm(a, m1, m2, n, max_iter, tol) | |
| # save results | |
| delta_0s.append(ksi_hat[0]) | |
| delta_1s.append(ksi_hat[1]) | |
| delta_2s.append(ksi_hat[2]) | |
| phi_10s.append(zeta_1_hat[0]) | |
| phi_11s.append(zeta_1_hat[1]) | |
| phi_12s.append(zeta_1_hat[2]) | |
| phi_20s.append(zeta_2_hat[0]) | |
| phi_21s.append(zeta_2_hat[1]) | |
| var_1s.append(var_1_hat) | |
| var_2s.append(var_2_hat) |
After running the simulations we check the results to confirm that the estimates are close to real values of the parameters.
On the screenshot above we can see that everything seems to work fine.
Now we are able to fit a model given its order, but how do we know which order to select? In the paper authors propose using Bayesian Information Criterion (or Schwarz Information Criterion). It is calculated as follows.
Note that the formula above is different from the one presented in the paper. The last term represents the number of parameters in the model. In the paper they start with generating three cointegrated time series and then use OLS to estimate the coefficients used to construct the spread. Therefore they have three parameters more to estimate. I start with directly generating a spread (time series following LMAR model), so I don’t need to estimate the cointegration coefficients and the total number of parameters is just equal to the sum of model dimensions m1+m2+n.
Now we need to compute the maximized log-likelihood Q. It is explained in the paper and I’ve described how to do it in the previous article, but I’m going to repeat it here briefly.
We have a mixture model, so we don’t really know from which regime a given datapoint comes. If we had that information (if we had a complete data), the log-likelihood would be computed as follows:
The variable Z above is an indicator of the current regime. These variables are not known, but we can replace them with their expected values - tau.
If we plug in tau in place of Z above, we will get the expected log-likelihood function Q. EM algorithm works by maximizing that function and we have everything we need to calculate it. I’m going to provide the code for the whole em_algorithm function again, but the only thing that changes is in the end (lines 132–137), where we calculate Q and return it along with the estimated parameters.
| def em_algorithm(a, m1, m2, n, max_iter, tol): | |
| ''' | |
| run em algorithm for max_iter iterations or until convergence | |
| a: time series | |
| m1: order of the first (stationary) AR model | |
| m2: order of the second (non-stationary) AR model | |
| n: number of lags in logistic equation | |
| ''' | |
| npdf = stats.norm.pdf | |
| max_lag = max(m1,m2,n) | |
| converged = False | |
| while not converged: | |
| #try: | |
| # random initial guess | |
| ksi = np.random.uniform(-3,3,(n+1)) | |
| zeta_1 = np.random.uniform(-3,3,(m1+1)) | |
| zeta_2 = np.random.uniform(-3,3,(m2+1)) | |
| var_1 = np.random.uniform(0,6,(1)) | |
| var_2 = np.random.uniform(0,6,(1)) | |
| # create dataframe | |
| a_lags = [f'a_l{i}' for i in range(1,max_lag+1)] | |
| abs_lags = [f'abs_l{i}' for i in range(1,n+1)] | |
| columns = ['const', 'a'] + a_lags + abs_lags + ['p1', 'p2', 'tau1', 'tau2', 'e1', 'e2'] | |
| df = pd.DataFrame(columns=columns) | |
| df['a'] = a[max_lag:] | |
| df['const'] = 1 | |
| for l in range(1,max_lag+1): | |
| df[a_lags[l-1]] = a[max_lag-l:-l] | |
| for l in range(1,n+1): | |
| df[abs_lags[l-1]] = abs(df[a_lags[l-1]]) | |
| for s in range(max_iter): | |
| # E-step | |
| # solve for mixing probabilities | |
| num = np.exp((df[['const'] + abs_lags] * ksi).sum(axis=1)) | |
| denom = (1 + np.exp((df[['const'] + abs_lags] * ksi).sum(axis=1))) | |
| df['p1'] = num/denom | |
| df['p2'] = 1 - df['p1'] | |
| # calculate e | |
| df['e1'] = df['a'] - (zeta_1 * df[['const'] + a_lags[:m1]]).sum(axis=1) | |
| df['e2'] = df['a'] - (zeta_2 * df[['const'] + a_lags[:m2]]).sum(axis=1) | |
| # calculate tau | |
| sigma_1 = np.sqrt(var_1) | |
| sigma_2 = np.sqrt(var_2) | |
| denom = ((df['p1'] / sigma_1) * stats.norm.pdf(df['e1'] / sigma_1) + | |
| (df['p2'] / sigma_2) * stats.norm.pdf(df['e2'] / sigma_2)) | |
| df['tau1'] = (df['p1'] / sigma_1) * stats.norm.pdf(df['e1'] / sigma_1) / denom | |
| df['tau2'] = (df['p2'] / sigma_2) * stats.norm.pdf(df['e2'] / sigma_2) / denom | |
| # M-step | |
| # compute ksi | |
| dr_dksi = df[['const'] + abs_lags] | |
| dQ_dksi = (np.tile((df['tau1'] - df['p1']), (n+1,1)).T * dr_dksi).sum().values | |
| df['dr_dksi^2'] = df[['const'] + abs_lags].apply(outer, axis=1).apply(np.array) | |
| d2Q_dksi2 = -(df['p1'] * df['p2'] * df['dr_dksi^2']).sum() | |
| new_ksi = ksi - np.linalg.inv(d2Q_dksi2) @ dQ_dksi | |
| # compute zeta | |
| df['de1_dzeta1^2'] = df[['const'] + a_lags[:m1]].apply(outer, axis=1).apply(np.array) | |
| df['de2_dzeta2^2'] = df[['const'] + a_lags[:m2]].apply(outer, axis=1).apply(np.array) | |
| zeta1_sum1 = (df['tau1'] * df['de1_dzeta1^2']).sum() | |
| zeta2_sum1 = (df['tau2'] * df['de2_dzeta2^2']).sum() | |
| zeta1_sum2 = (-np.tile(df['tau1'] * df['a'], (m1+1,1)).T * -df[['const'] + a_lags[:m1]]).sum().values | |
| zeta2_sum2 = (-np.tile(df['tau2'] * df['a'], (m2+1,1)).T * -df[['const'] + a_lags[:m2]]).sum().values | |
| new_zeta_1 = np.linalg.inv(zeta1_sum1) @ zeta1_sum2 | |
| new_zeta_2 = np.linalg.inv(zeta2_sum1) @ zeta2_sum2 | |
| # compute variance | |
| sigma1_sum2 = (df['tau1'] * (df['a'] + np.dot(-df[['const'] + a_lags[:m1]], new_zeta_1))**2).sum() | |
| sigma2_sum2 = (df['tau2'] * (df['a'] + np.dot(-df[['const'] + a_lags[:m2]], new_zeta_2))**2).sum() | |
| new_var_1 = 1/df['tau1'].sum() * sigma1_sum2 | |
| new_var_2 = 1/df['tau2'].sum() * sigma2_sum2 | |
| # calculate the difference between old and new values of parameters | |
| delta_ksi = np.sum(abs(new_ksi - ksi)) | |
| delta_zeta_1 = np.sum(abs(new_zeta_1 - zeta_1)) | |
| delta_zeta_2 = np.sum(abs(new_zeta_2 - zeta_2)) | |
| delta_var_1 = np.sum(abs(new_var_1 - var_1)) | |
| delta_var_2 = np.sum(abs(new_var_2 - var_2)) | |
| # update parameters | |
| ksi = new_ksi | |
| zeta_1 = new_zeta_1 | |
| zeta_2 = new_zeta_2 | |
| var_1 = new_var_1 | |
| var_2 = new_var_2 | |
| # if any parameter is nan, stop | |
| nansum = (np.isnan(ksi).sum() + np.isnan(zeta_1).sum() + np.isnan(zeta_2).sum() + | |
| np.isnan(var_1).sum() + np.isnan(var_2).sum()) | |
| if nansum != 0: | |
| break | |
| # calculate total difference between old and new parameters | |
| delta_total = delta_ksi + delta_zeta_1 + delta_zeta_2 + delta_var_1 + delta_var_2 | |
| # check convergence | |
| if delta_total < tol: | |
| # calculate roots of characteristic polynomial of the second regime | |
| char_pol = np.polynomial.polynomial.Polynomial(np.concatenate(([1],-zeta_2[1:]))) | |
| # calculate reciprocal roots and take absolute value | |
| roots = np.abs(1/char_pol.roots()) | |
| # convergence conditions | |
| # second regime is non-stationary | |
| conv_cond = sum(roots>=1) > 0 # at least one reciprocal root >= 1 | |
| if conv_cond: | |
| print('Converged at iteration ', s) | |
| converged = True | |
| break | |
| else: | |
| break | |
| if not converged: | |
| print(f'Algorithm not converged') | |
| print('Repeating with different initital values') | |
| #except: | |
| # continue | |
| # calculate Q | |
| Q_sum1 = (df['tau1'] * (np.log(df['p1']) - 1/2*np.log(var_1) - df['e1']**2 / (2 * var_1))).sum() | |
| Q_sum2 = (df['tau2'] * (np.log(df['p2']) - 1/2*np.log(var_2) - df['e2']**2 / (2 * var_2))).sum() | |
| Q = Q_sum1 + Q_sum2 | |
| return ksi, zeta_1, var_1, zeta_2, var_2, Q |
With that function we can perform simulation studies of model selection. These simulations are even more time consuming than the previous ones, so I’m not going to perform as many simulations as in the paper and I will only use two possible values for each of the parameters (m1=1,2, m2=1,2, n=1,2). This means that we will try to fit 8 different models at each simulation and select the one with the smallest BIC. Then we will see what percentage of the simulations correctly identify the model dimensions.
Code for running such simulations is shown below.
| import warnings | |
| import itertools | |
| bic_df = pd.DataFrame(columns=['m1', 'm2', 'n', 'BIC']) | |
| max_iter=50 | |
| tol = 0.05 | |
| N = 1000 # number of datapoints to generate | |
| N_sim = 50 # number of simulations to run | |
| # parameters used to generate a | |
| ksi = [-1.3, 0.6, 0.3] | |
| zeta_1 = [0, 0.6, -0.2] | |
| var_1 = 1 | |
| zeta_2 = [0, 1.5] | |
| var_2 = 3 | |
| for s in range(N_sim): | |
| print('Running sim ', s) | |
| # generate a | |
| np.random.seed(123+s) | |
| a = generate_a(N, ksi, zeta_1, var_1, zeta_2, var_2) | |
| best_bic = np.inf | |
| best_m1, best_m2, best_n = 0,0,0 | |
| for m1,m2,n in itertools.product([1,2],[1,2],[1,2]): | |
| with warnings.catch_warnings(): | |
| warnings.simplefilter('ignore') # supress warnings from EM algorithm | |
| # run EM algorithm to estimate parameters | |
| ksi_hat, zeta_1_hat, var_1_hat, zeta_2_hat, var_2_hat, Q = em_algorithm(a, m1, m2, n, max_iter, tol) | |
| # compute bic | |
| bic = -2*Q + np.log(N - max(m1,m2,n)) * (m1 + m2 + n) | |
| if bic < best_bic: | |
| best_bic = bic | |
| best_m1, best_m2, best_n = m1,m2,n | |
| print(f'm1={best_m1}, m2={best_m2}, n={best_n}, BIC={best_bic}') | |
| bic_df.loc[s] = best_m1, best_m2, best_n, best_bic | |
| print() |
First several lines of the resulting dataframe are demonstrated on the screenshot below.
Now let’s see in what percentage of the simulations each individual dimension was identified correctly and in what percentage of the simulations all of the dimensions were identified correctly.
8% of the simulations identified parameter m1 correctly. 48% identified parameter m2 correctly. 40% identified parameter n correctly. And only 22% of the simulations were able to correctly identify all the dimensions.
These numbers are lower than the ones provided in the paper, but probably it’s just a statistical fluke because I didn't run enough simulations (only 50 versus 300 in the paper). Also we need to keep in mind that when applied to real world data no model would be an ideal fit, we just need to identify one that is good enough.
Now that we have a working implementation of the EM algorithm, we can try to implement a trading strategy based on LMAR model, but I’ll leave it for another article.
If you have any questions, suggestions or corrections please post them in the comments. Thanks for reading.
References
[1] Basket trading under co-integration with the logistic mixture autoregressive model
[2] On a logistic mixture autoregressive model
[3] https://en.wikipedia.org/wiki/Identifiability
[4] https://en.wikipedia.org/wiki/Bayesian_information_criterion